Antifungal resistance in pathogenic fungi is a growing global health concern. Non-pathogenic Saccharomyces cerevisiae is a useful model for studying mechanisms of antifungal resistance that are relevant to understanding the same processes in pathogenic fungi. yEvo high school students used experimental evolution to study antifungal resistance by growing yeast in increasing concentrations of different types of antifungal drugs, including this project on azoles and our other projects on echinocandins.
Results
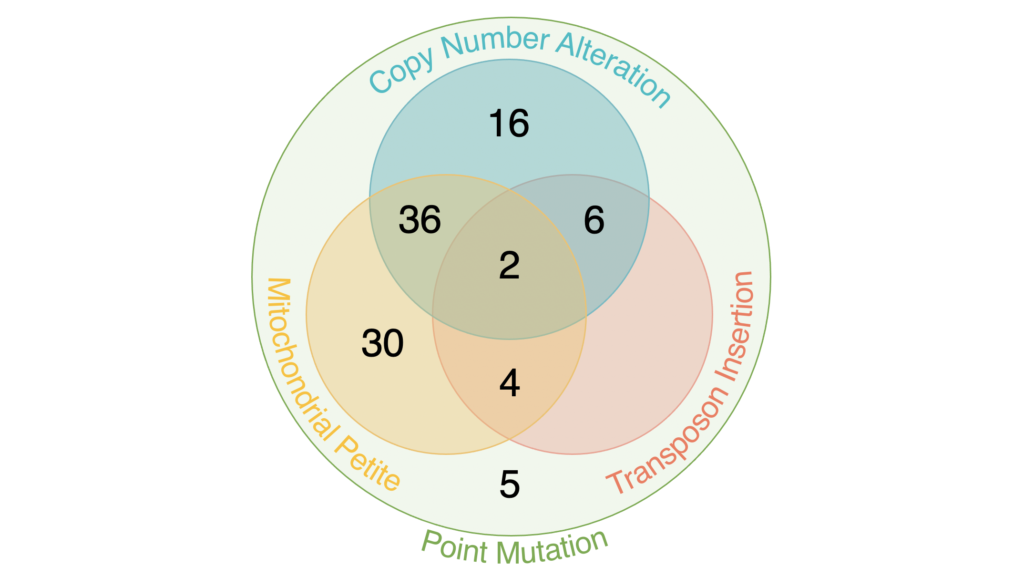
Our university labs sequenced the DNA from 99 azole-resistant clones and found mutations previously shown to impact azole resistance, demonstrating the efficacy of our protocols. We found each evolved strain had at least one point mutation, and many had copy number alterations or mitochondrial genome loss. A few had transposon mobilization events.
Many of the mutations were in genes with functions known to be involved in azole resistance: in drug efflux, and in sterol and sphingolipid synthesis. These findings helped validate our protocol for experimental evolution by students.
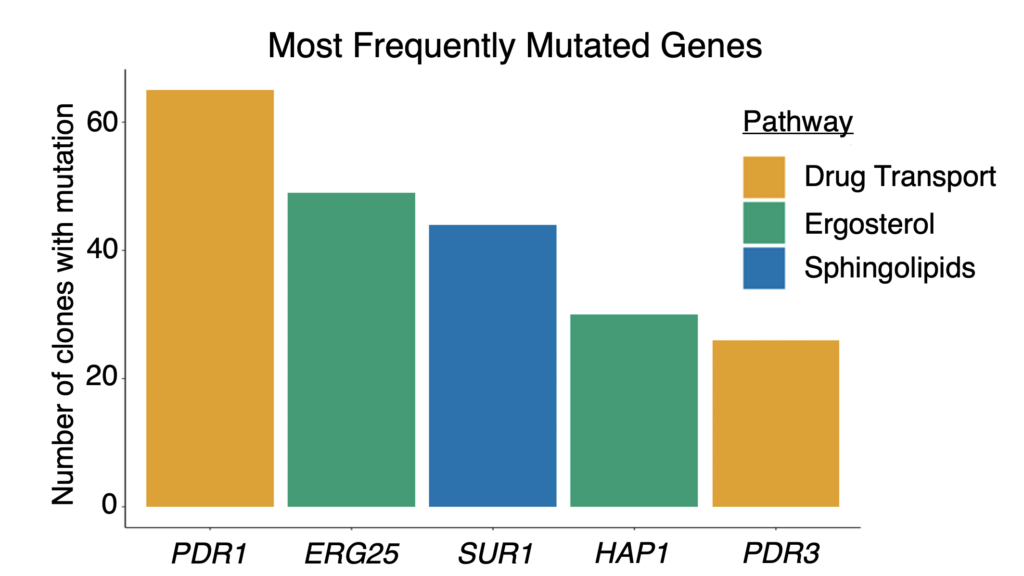
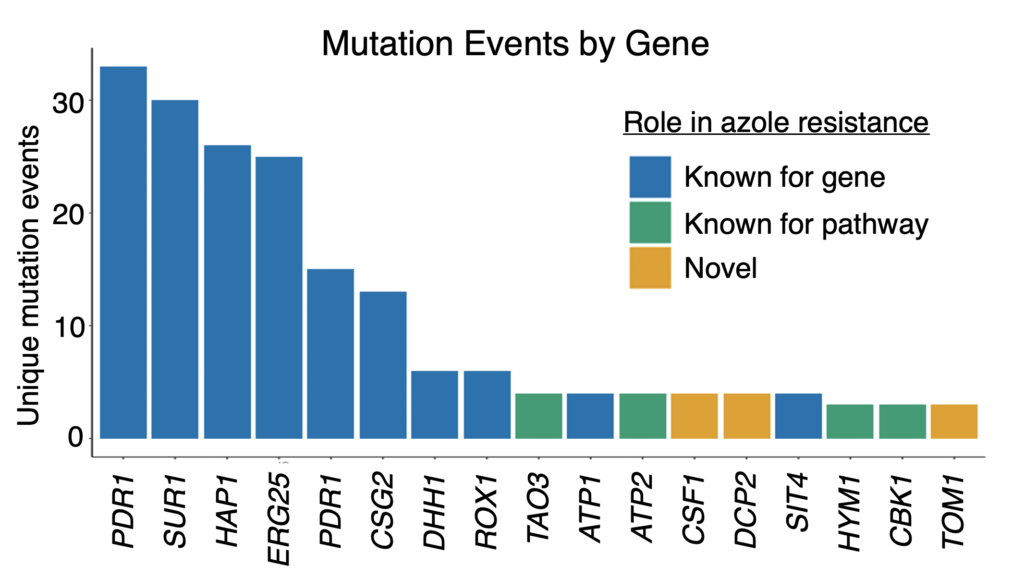
We also found mutations in genes not previously implicated in azole resistance. Some were in pathways that affect resistance, such as RAM signaling. We additionally found recurrent mutations in an mRNA degradation pathway and an uncharacterized mitochondrial protein (Csf1) that have possible novel mechanistic connections to azole resistance.
The scale of replication also allowed us to identify epistatic interactions, as evidenced by pairs of mutations that occur in the same clone more frequently than expected by chance (positive epistasis) or less frequently (negative epistasis). We validated one of these pairs, a negative epistatic interaction between gain-of-function mutations in the multidrug resistance transcription factors Pdr1 and Pdr3. This project established that yEvo’s high school-university collaboration can serve as a model for involving members of the broader science-interested public in the scientific process to make meaningful discoveries in biomedical research.
Read more about our results from our publications!
Resources
We have developed a series of modules that can be used to explore azole resistance in yeast; you can view our general protocol and specific modules.
We also developed a Yeast Madness kit to compare the fitness of different azole-resistant evolved yeast in collaboration with Fred Hutch Science Education Project. Resources and protocol are available from Fred Hutch, kits can be requested but must be picked up in Seattle, WA.
Participants
This project was carried out with high school educators at Westridge School and Moscow High School, the Dunham Lab at University of Washington, and the Rowley Lab at the University of Idaho.
