You have sequencing data from your evolved yeast - now what?
We have modules where you can learn how to:
- Align sequencing reads to a reference genome to identify differences between your evolved yeast and the reference yeast genome sequence
- Visualize sequencing read alignments to determine which differences are unique to your evolved yeast
Part 1: Align sequencing data
You can use Snippy on Galaxy Europe to map your sequencing reads to the reference yeast genome.
Download our worksheet for Aligning Reads Using Snippy. You will need .fastq sequencing files; many are available from NCBI for published datasets including yEvo azole evolutions (Taylor et al. 2022).
This module is still in development. If you have feedback, especially for ways to compare the evolved and ancestor yeast, please reach out to the Dunham lab!
Part 2: Visualize sequencing data
You can use the IGV web app to visualize sequencing reads and identify mutations.
Download our worksheet for Viewing Mutations in Sequencing Data and the accompanying tutorial data (follow link, then click "Download full dataset" on the right sidebar). You can also use this to analyze files generated by Snippy in Part 1.
In this lesson, you will learn to:
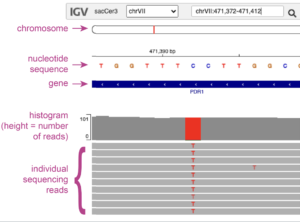
Evaluate potential mutations
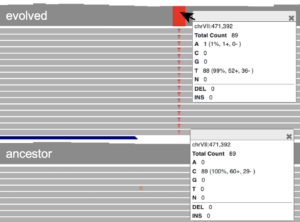
Explore mutation types
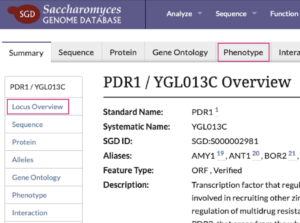
Learn about affected genes
